Results
Puzzle 34
Sequence (5' to 3'):
GCGGGCUGACCGACCCCCCGAGUUCGCUCGGGGACAACUAGA CAUACAGUAUGAAAAUACUGAGCCCGC
Puzzle: methyl transferase ribozyme
Crystal structure kindly provided by: Lin Huang
Reference: Structure and mechanism of a methyltransferase ribozyme. Nat Chem Biol. (2022)
PDB id: 7v9e
Puzzle 33

Sequence (5' to 3'):
GGAGUAGAAGCGUUCAGCGGCCGAAAGGCCGCCCGGAAAUUGCUCC
Puzzle: xanthine riboswitch (Ideonella sp. B508-1)
Crystal structure kindly provided by: Aiming Ren
Reference: Insights into xanthine riboswitch structure and metal ion-mediated ligand recognition. Nucleic Acids Res (2021)
Puzzle 32
Sequence (5' to 3'):
GGCGCACUGGCGCUGCGCCUUCGGGCGCCAAUCGUAGCGUGU CGGCGCC
Puzzle: Pepper Aptamer
Crystal structure kindly provided by: Aiming Ren
Reference: Structure-based investigation of fluorogenic Pepper aptamer. Nat Chem Biol (2021) 17: 1289-1295
PDB id: 7eoj
Puzzle 31
Sequence (5' to 3'):
GGCGGUGUAAGUGCAGCCCGUCUUACACCGUGCGGCACAGAA ACACUGAUGUCGUAUACAGGGCG
Puzzle: PFSE in Sars-CoV-2
Crystal structure kindly provided by: Joseph A Piccirilli
Reference: The SARS-CoV-2 Programmed -1 Ribosomal Frameshifting Element Crystal Structure Solved to 2.09 angstrom Using Chaperone-Assisted RNA Crystallography. ACS Chem Biol (2021) 16: 1469-1481
PDB id: 7mlx
Puzzle 30
Sequence (5' to 3'):
GAACCCCGCCUGGAGGCCGCGGUCGGCCCGGGGCUUCUCCGG AGGCACCCACUGCCACCGCGAAGAGUUGGGCUCUGUCAGCCG CGGG
Puzzle: human telomerase RNA
Crystal structure kindly provided by: Thi Hoang Duong Nguyen
Reference: Structure of human telomerase holoenzyme with bound telomeric DNA. Nature (2021) 593: 449-453
PDB id: 7bg9
Puzzle 29
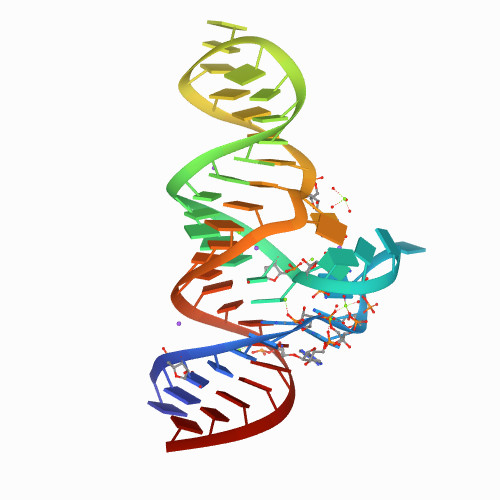
Sequence (5' to 3'):
GGCUUCAACAACCCCGUAGGUUGGGCCGAAAGGCAGCGAAUCUACUGGAGCC
Puzzle: NAD+ riboswitch (Candidatus Koribacter versatilis Ellin345)
Crystal structure kindly provided by: Lin Huang
Reference: Structure and ligand binding of the ADP-binding domain of the NAD + riboswitch. RNA (2020) 26: 878-887
PDB id: 6tb7
Puzzle 28
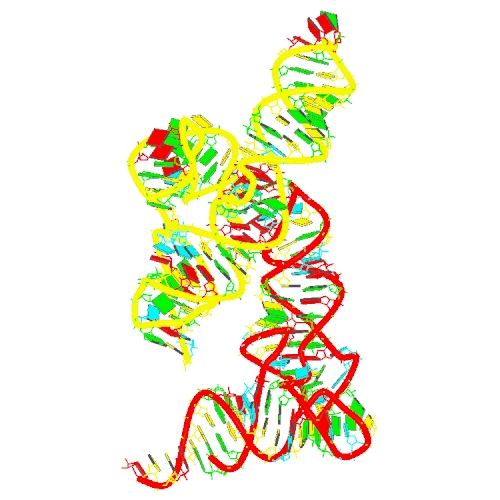
Sequence (5' to 3'):
tRNA:
GGGCCUAUAGCUCAGGCGGUUAGAGCGCUUCGCUGAUAACGA
AGAGGUCGGAGGUUCGAGUCCUCCUAGGCCCACCA
T-box:
GGCGACGAUCCGGCCAUCACCGGGGAGCCUUCGGAAGAACGG
CGCCGCCGGAAACGGCGGCGCUCAGUAGAACCGAACGGGUGA
GCCCGUCACAGCUC
Puzzle: T-box riboswitch - tRNA complex (Nocardia farcinica ileS)
Crystal structure kindly provided by: Jingwei Zhang
Reference: High-affinity recognition of specific tRNAs by an mRNA anticodon-binding groove. Nat Struct Mol Biol. 2019 Dec;26(12):1114-1122. doi: 10.1038/s41594-019-0335-6. Epub 2019 Dec 2.
PDB id: 6ufm
raw prediction | Assessment results Complex T-box
Puzzle 27
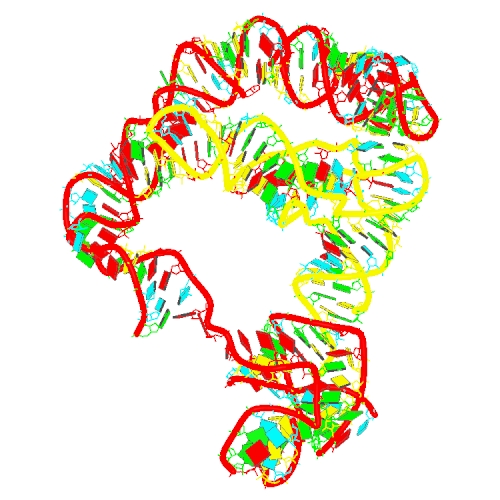
Sequence (5' to 3'):
tRNA:
GCGGAAGTAGTTCAGTGGTAGAACACCACCTTGCCAAGGTGGG
GGTCGCGGGTTCGAATCCCGTCTTCCGCTCCA
T-box:
GUUGCAGUGAGAGAAAGAAGUACUUGCGUUUACCUCAUGAAAG
CGACCUUAGGGCGGUGUAAGCUAAGGAUGAGCACGCAACGAAA
GGCAUUCUUGAGCAAUUUUAAAAAAGAGGCUGGGAUUUUGUUC
UCAGCAACUAGGGUGGAACCGCGGGAGAACUCUCGUCCCUA
Puzzle: T-box riboswitch - tRNA complex (Bacillus subtilis glyQS)
Crystal structure kindly provided by: Jingwei Zhang
Reference: Structural basis of amino acid surveillance by higher-order tRNA-mRNA interactions. Nat Struct Mol Biol. 2019 Dec;26(12):1094-1105. doi: 10.1038/s41594-019-0326-7. Epub 2019 Nov 18.
PDB id: 6pom
raw prediction | Assessment results Complex T-box
Puzzle 26
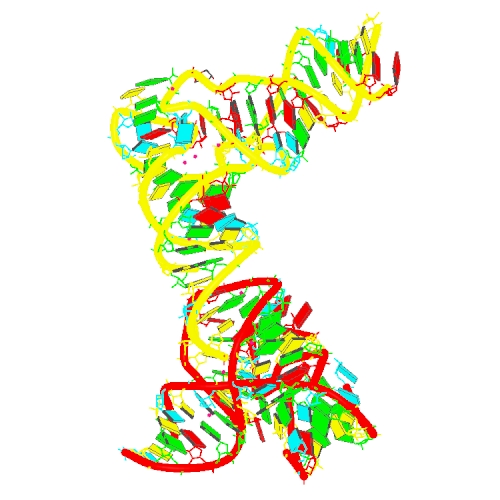
Sequence (5' to 3'):
T-box:
GCGGAAGTAGTTCAGTGGTAGAACACCACCTTGCCAAGGTGG
GGGTCGCGGGTTCGAATCCCGTCTTCCGCTCCA
tRNA:
GAAAGUGGGUGCGCGUUUGGCGCAUCAACUCGGGUGGAACCG
CGGGAGCUACGCUCUCGUCCCGAG
Puzzle: T-box riboswitch - tRNA complex (Geobacillus kaustophilus glyQ)
Crystal structure kindly provided by: Jingwei Zhang
Reference: Structural basis of amino acid surveillance by higher-order tRNA-mRNA interactions. Nat Struct Mol Biol. 2019 Dec;26(12):1094-1105. doi: 10.1038/s41594-019-0326-7. Epub 2019 Nov 18.
PDB id: 6pmo
raw prediction | Assessment results Complex T-box
Puzzle 25
Sequence (5' to 3'):
GGGUGUAAUCUCCAAAAUAUGGUUGGGGAGCCUCCACCAGUGAA CCGUAAAAUCGCUGUCACCACCCAG
Puzzle: 2'-dG-II riboswitch
Crystal structure kindly provided by: Batey, R.T.
Reference: Structural basis for 2'-deoxyguanosine recognition by the 2'-dG-II class of riboswitches. Nucleic Acids Res. 2019 Nov 18;47(20):10931-10941. doi: 10.1093/nar/gkz839.
PDB id: 6p2h
Puzzle 24
Sequence (5' to 3'):
GGACCUCGCAAGGGUAUCAUGGCGGACGACCGGGGUUCGAACCCCGGAUC
CGGCCGUCCGCCGUGAUCCAUGCGGUUACCGCCCGCGUGUCGAACCCAGG
UGUGCGAGGUCC
Puzzle: adenovirus virus-associated RNA
Crystal structure kindly provided by: Jingwei Zhang
Reference: Crystal structure of an adenovirus virus-associated RNA. Nature Communicationsvolume 10, Article number: 2871 (2019).
PDB id: 6ol3
Puzzle 23
Sequence (5' to 3'):
GUACGAAGGAAGGUUUGGUAUGGGGUAGUUGUCGUAC
Puzzle: Mango-III aptamer
Crystal structure kindly provided by: Ferre-D'Amare, A.R.
Reference: Structure and functional reselection of the Mango-III fluorogenic RNA aptamer. Nat Chem Biol. 2019 May;15(5):472-479. doi: 10.1038/s41589-019-0267-9. Epub 2019 Apr 15.
PDB id: 6e8u
Puzzle 22
Sequence (5' to 3'):
UUACUGUGAGAAUCAGUAACAAACAUGUGGGGCUUAUAU CUAAUCGAAAGAUUAGUAUUAGUGCAGAC
GUUAAAACCAUGUC
Puzzle: Hatchet Ribozyme
Crystal structure kindly provided by: Aiming Ren
Reference: Hatchet ribozyme structure and implications for cleavage mechanism. Proc Natl Acad Sci U S A. 2019 May 28;116(22):10783-10791. doi: 10.1073/pnas.1902413116. Epub 2019 May 14.
raw prediction | Assessment results monomer dimer
Puzzle 21
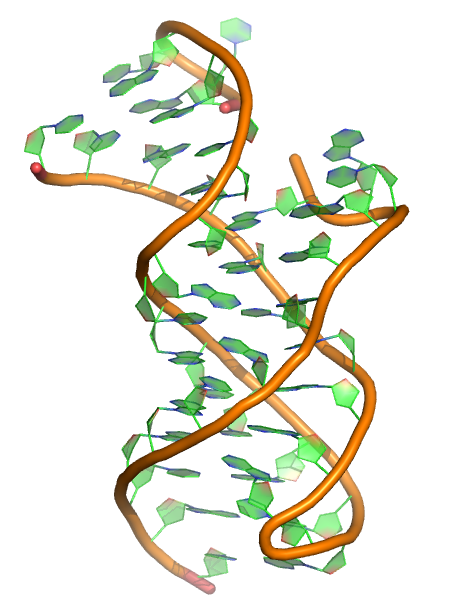
Sequence (5' to 3'):
CCGGACGAGGUGCGCCGUACCCGGUCAGGACAAGACGGCGC
Puzzle: Guanidine III Riboswitch
Crystal structure kindly provided by: David M J Lilley
Reference: Structure of the Guanidine III Riboswitch. Cell Chem Biol. 2017 Nov 16;24(11):1407-1415.e2. doi: 10.1016/j.chembiol.2017.08.021. Epub 2017 Oct 5.
Puzzle 20
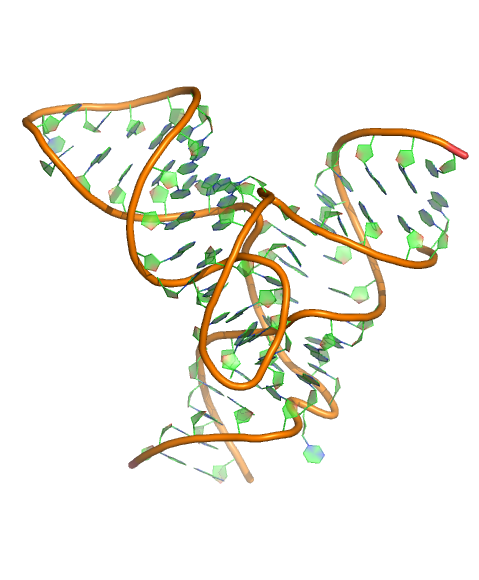
Sequence (5' to 3'):
ACCCGCAAGGCCGACGGC-GCCGCCGCUGGUGCAAGUCCAGCCACGCUUC
GGCGUGGGCGCUCAUGGGU
Puzzle: Another Twister Sister ribozyme
Crystal structure kindly provided by: Amy Ren
Reference: Zheng, L., Mairhofer, E., Teplova, M., Zhang, Y., Ma, J., Patel, D.J., Micura, R., Ren, A. Structure-based insights into self-cleavage by a four-way junctional twister-sister ribozyme. (2017) Nat Commun 8: 1180-1180
Puzzle 19
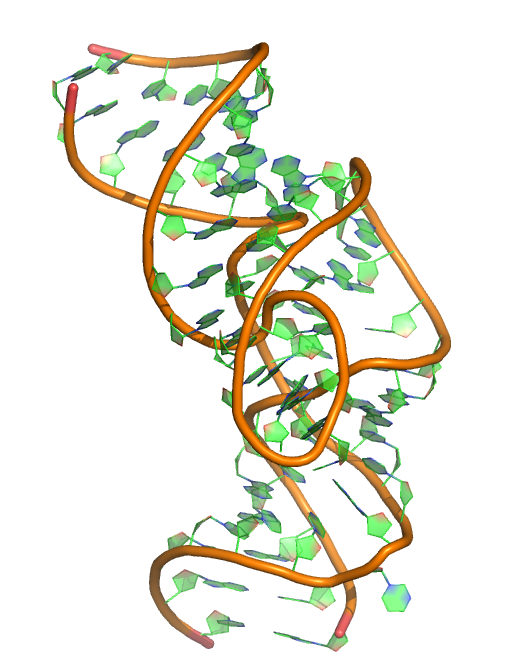
Sequence (5' to 3'):
GCAGGGCAAGGCCCAGUCCCGUGCAAGCCGGGACCGCCCC- GGGGCGCGGCGCUCAUUCCUGC
Puzzle: One Twister Sister robozyme
Crystal structure kindly provided by: David M J Lilley
Reference: The structure of a nucleolytic ribozyme that employs a catalytic metal ion. Nature Chemical Biology 13, 508–513 (2017) doi:10.1038/nchembio.2333.
PDB id: 5t5a
Puzzle 18
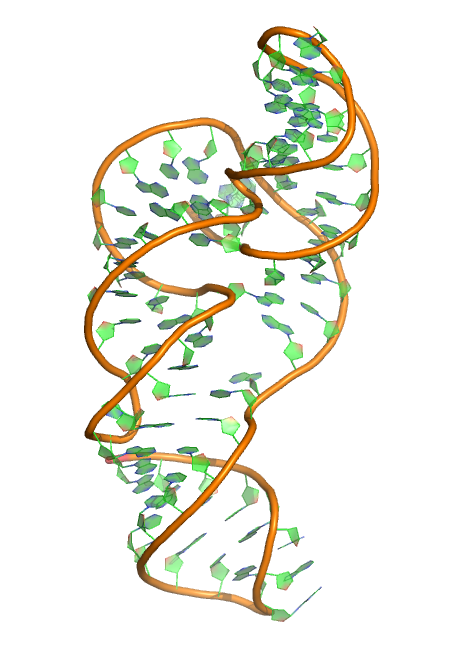
Sequence (5' to 3'): GGGUCAGGCCGGCGAAAGUCGCCACAGUUUGGGGAAAGCUGUGCAGCCUG UAACCCCCCCACGAAAGUGGG
Puzzle: Zika virus.
Crystal structure kindly provided by: Jeffrey S. Kieft
Reference: Zika virus produces noncoding RNAs using a multi-pseudoknot structure that confounds a cellular exonuclease. (2016) Science 354: 1148-1152. DOI: 10.1126/science.aah3963.
PDB id: 5tpy
Puzzle 17
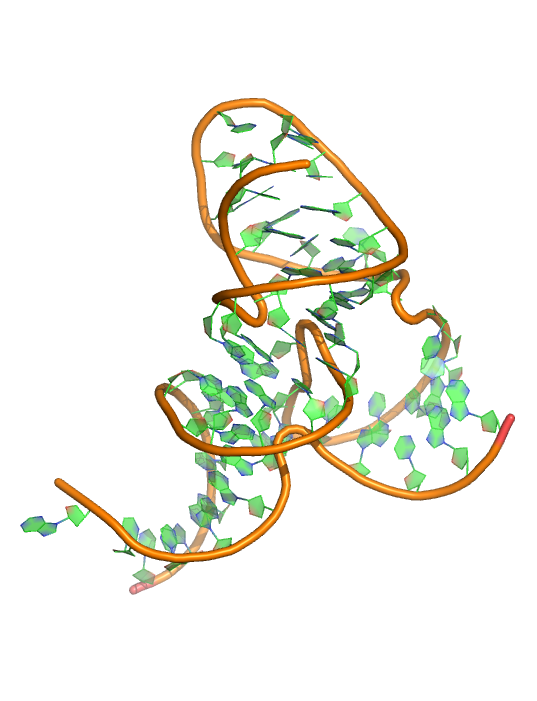
Sequence (5' to 3'): CGUGGUUAGGGCCACGUUAAAUAGUUGCUUAAGCCCUAAGCGUUGAUAAA UAUCAGGUGCAA
Puzzle: Pistol ribozyme.
Crystal structure kindly provided by: Dinshaw Patel
Reference: Pistol ribozyme adopts a pseudoknot fold facilitating site-specific in-line cleavage. Nature Chemical Biology 12, 702–708 (2016) doi:10.1038/nchembio.2125.
PDB id: 5k7c
Puzzle 16

Sequence (5' to 3'):
seq1: UGCUCCUAGUACGAGAGGAACGGAGUG
seq2: UGCUCCGAGUACGAGAGGAAAGGAGUG
Puzzle: Sarcin Ricin Loop
Crystal structure kindly provided by: Eric Ennifar
Reference: To be published.
raw prediction seq1 raw prediction seq2 | Assessment results seq1 Assessment results seq2
Puzzle 15
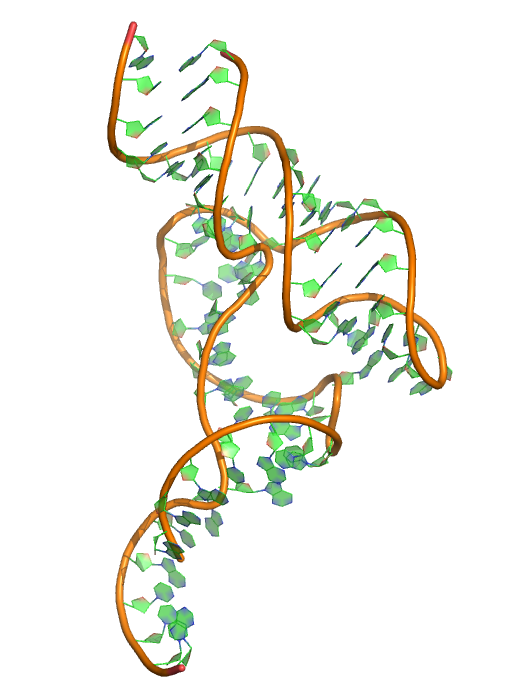
Sequence (5' to 3'): GGGUACUUAAGCCCACUGAUGAGUCGCUGGGAUGCGACGAAACGCCCA- GGGCGUCUGGGCAGUACCCA
Puzzle: Hammerhead Ribozyme. Predict as 2 chains.
Crystal structure kindly provided by: Barbara L. Golden
Reference: Two Active Site Divalent Ions in the Crystal Structure of the Hammerhead Ribozyme Bound to a Transition State Analogue. Biochemistry, 2016, 55 (4), pp 633–636 DOI: 10.1021/acs.biochem.5b01139.
PDB id: 5di4
Puzzle 14
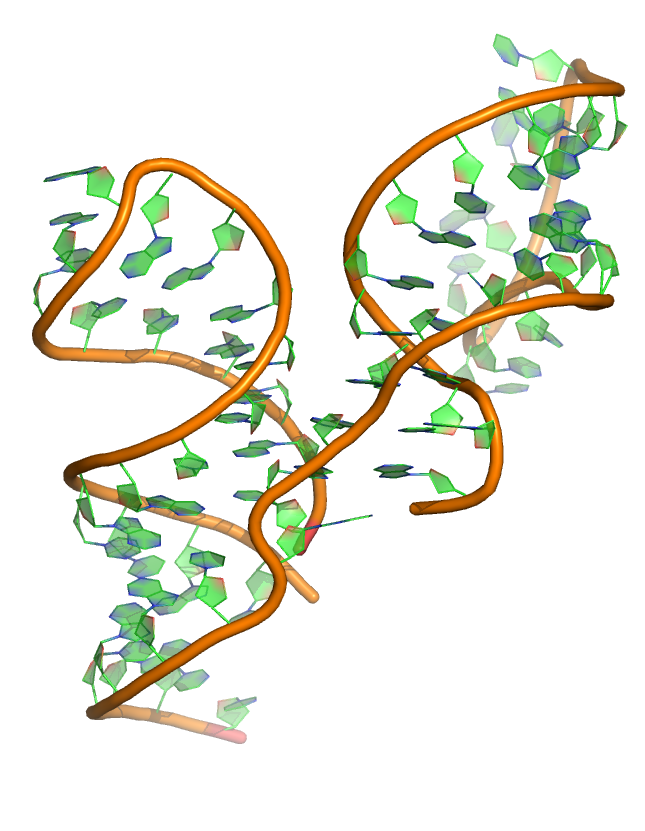
Sequence (5' to 3'):
Bound form:
CGUUGACCCAGGAAACUGGGCGGAAGUAAGGCCCAUUGCACUCCGGGCCUGAAG
CAACGCG
Free form:
CGUUGGCCCAGGAAACUGGGUGGAAGUAAGGCCCAUUGCACUCCGGGCCUGAAG
CAACGCU
Puzzle: L-glutamine riboswitch. Predict both Free and Bound forms.
Crystal structure kindly provided by: Dinshaw Patel
Reference: Aiming Ren, Yi Xue, Alla Peselis, Alexander Serganov, Hashim M. Al-Hashimi, Dinshaw J. Patel. Structural and Dynamic Basis for Low-Affinity, High- Selectivity Binding of L-Glutamine by the Glutamine Riboswitch. Cell Rep. 2015 Dec 1;13(9):1800-13.01251-6)
PDB id: 5ddo
Bound Form: raw prediction | Assessment results
Free Form: raw prediction | Assessment results
Puzzle 13
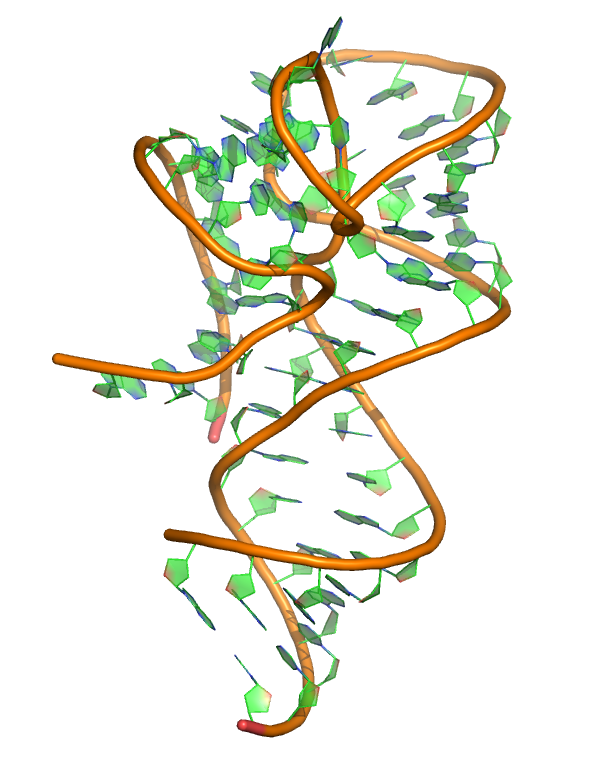
Sequence (5' to 3'): GGGUCGUGACUGGCGAACAGGUGGGAAACCACCGGGGAGCGACCCCGGCAUCGA UAGCCGCCCGCCUGGGC
Puzzle: ZMP Riboswitch.
Crystal structure kindly provided by: Robert T. Batey
Reference: Trausch JJ, Marcano-Velázquez JG, Matyjasik MM, Batey RT. Metal Ion-Mediated Nucleobase Recognition by the ZTP Riboswitch. Chem Biol. 2015 Jul 23;22(7):829-37.
PDB id: 4xw7
Puzzle 12
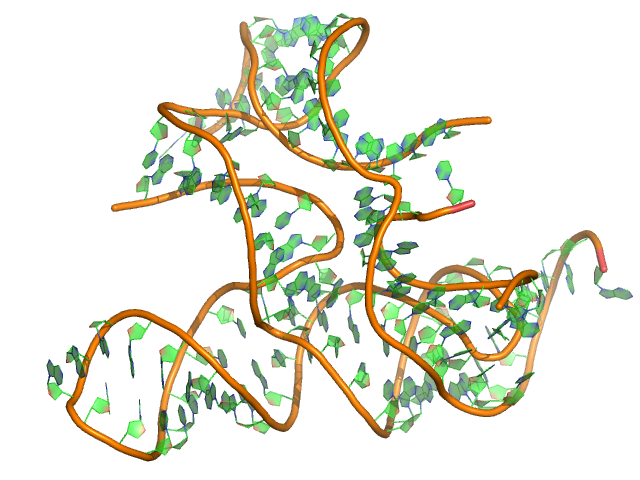
Sequence (5' to 3'): AUCGCUGAACGCGGGGGACCCAGGGGGCGAAUCUCUUCCGAAAGGAAGAGUAGG GUUACUCCUUCGACCCGAGCCCGUCAGCUAACCUCGCAAGCGUCCGAAGGAGAA AAA
Puzzle: ydaO riboswitch.
Crystal structure kindly provided by: Dinshaw Patel
Reference: Ren A, Patel DJ. 2014. c-di-AMP binds the ydaO riboswitch in two pseudo-symmetry-related pockets. Nat Chem Biol. 10(9):780-6.
PDB id: 4qlm
Puzzle 11
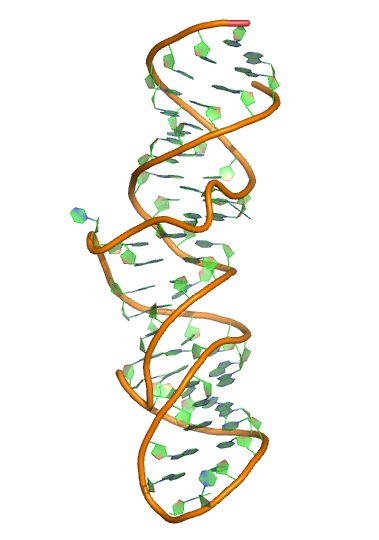
Sequence (5' to 3'):
GGGAUCUGUCACCCCAUUGAUCGCCUUCGGGCUGAUCUGGCUGG
CUAGGCGGGUCCC
Puzzle: 7SK
Crystal structure kindly provided by: Anne-Catherine Dock-Bregeon
Reference: Martinez-Zapien D, Legrand P, McEwen AG, Proux F, Cragnolini T, Pasquali S, Dock-Bregeon AC. The crystal structure of the 5΄ functional domain of the transcription riboregulator 7SK. Nucleic Acids Res. 2017 Apr 7;45(6):3568-3579. doi: 10.1093/nar/gkw1351. Bourbigot S, Dock-Bregeon AC, Eberling P, Coutant J, Kieffer B, Lebars I. Solution structure of the 5'-terminal hairpin of the 7SK small nuclear RNA. RNA. 2016 Dec;22(12):1844-1858. Epub 2016 Oct 20.
Puzzle 10
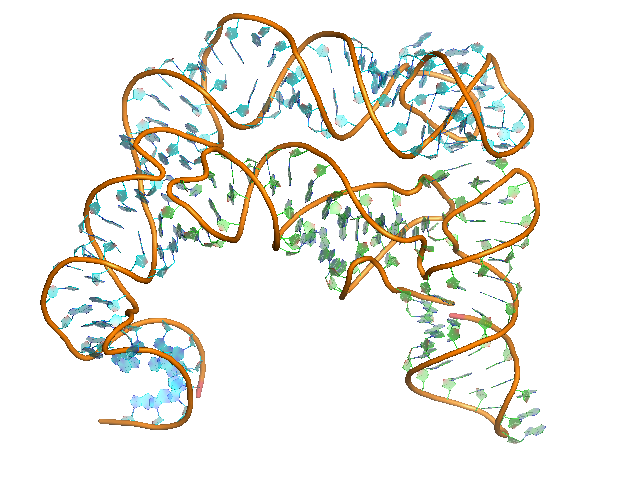
Sequence (5' to 3'):
T-box:
UGCGAUGAGAAGAAGAGUAUUAAGGAUUUACUAUGAUUAGCGACUCUAGGAUAG
UGAAAGCUAGAGGAUAGUAACCUUAAGAAGGCACUUCGAGCA
tRNA:
GCGGAAGUAGUUCAGUGGUAGAACACCACCUUGCCAAGGUGGGGGUCGCGGGUU
CGAAUCCCGUCUUCCGCUCCA
Puzzle: T-box riboswitch - tRNA complex.
Crystal structure kindly provided by: Adrian Ferré-D'amaré
Reference: Zhang, J. & Ferré-D'Amaré, A.R. Co-crystal structure of a T-box riboswitch stem I domain in complex with its cognate tRNA. Nature 500, 363-366 (2013).
PDB id: 4lck
Assessment of complex: raw prediction | Assessment results
Assessment of tBox: raw prediction | Assessment results
Assessment of tBox: raw prediction | Assessment results
Puzzle 9
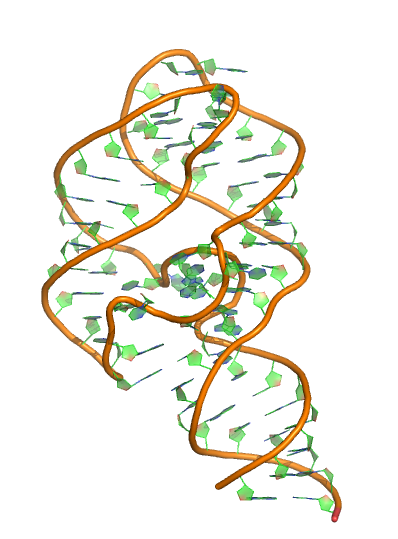
Sequence (5' to 3'):
GGACACUGAUGAUCGCGUGGAUAUGGCACGCAUUGAAUUGU
UGGACACCGUAAAUGUCCUAACACGUGUCC
Puzzle: 5-hydroxytryptophan aptamer
Crystal structure kindly provided by: Robert T. Batey
Reference: Ely B Porter, Jacob T Polaski, Makenna M Morck & Robert T Batey. Recurrent RNA motifs as scaffolds for genetically encodable small-molecule biosensors. Nature Chemical Biology volume 13, pages 295–301 (2017)
PDB id: 5kpy
Puzzle 8
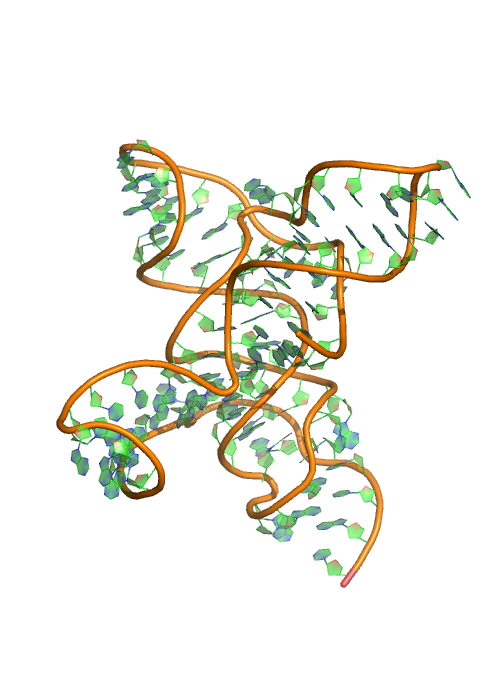
Sequence (5' to 3'): GGAUCACGAGGGGGAGACCCCGGCAACCUGGGACGGACACCCAAGGUGCUCACA CCGGAGACGGUGGAUCCGGCCCGAGAGGGCAACGAAGUCCGU
Puzzle: SAM riboswitch.
Crystal structure kindly provided by: Robert T. Batey
Reference: Trausch JJ, Xu Z, Edwards AL, Reyes FE, Ross PE, Knight R, Batey RT. 2014. Structural basis for diversity in the SAM clan of riboswitches. PNAS.111(18):6624-9.
PDB id: 4l81
Puzzle 7
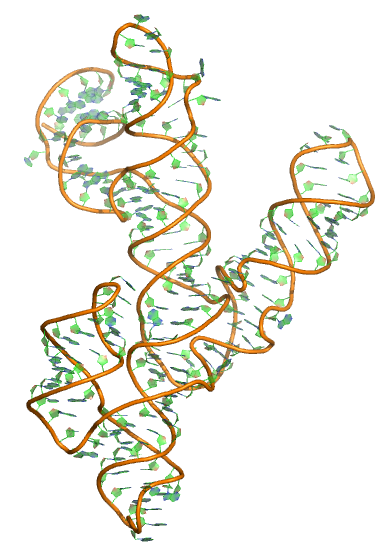
Sequence (5' to 3'): GCGCUGUGUCGCAAUCUGCGAAGGGCGUCGUCGGCCCGAGCGGUAGUAAGCAGG GAACUCACCUCCAAUGAAACACAUUGUCGUAGCAGUUGACUACUGUUAUGUGAU UGGUAGAGGCUAAGUGACGGUAUUGGCGUAAGCCAAUACCGCGGCACAGCACAA GCCCGCUUGCGAGAUUACAGCGC
Puzzle: Varkud satellite ribozyme.
Crystal structure kindly provided by: Joseph A Piccirilli
Reference: Nikolai B Suslov, Saurja DasGupta, Hao Huang, James R Fuller, David M J Lilley, Phoebe A Rice& Joseph A Piccirilli. 2015. Crystal structure of the Varkud satellite ribozyme. Nature Chemical Biology 11, 840–846.
PDB id: 4r4v
Puzzle 6
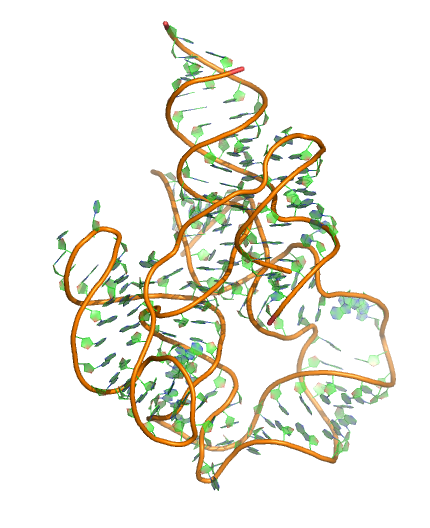
Sequence (5' to 3'): CGGCAGGUGCUCCCGACCCUGCGGUCGGGAGUUAAAAGGGAAGCCGGUGCAAGU CCGGCACGGUCCCGCCACUGUGACGGGGAGUCGCCCCUCGGGAUGUGCCACUGG CCCGAAGGCCGGGAAGGCGGAGGGGCGGCGAGGAUCCGGAGUCAGGAAACCUGC CUGCCG
Puzzle: adenosylcobalamin riboswitch.
Crystal structure kindly provided by: Alexnder Serganov
Reference: Peselis A and Serganov A. 2012. Structural insights into ligand binding and gene expression control by an adenosylcobalamin riboswitch. Nature Structural and Molecular Biology 19 (11), 1182-1184.
PDB id: 4gxy
Puzzle 5
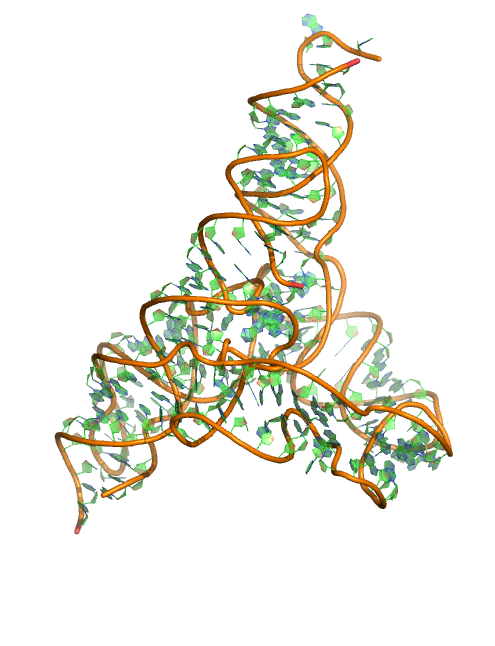
Sequence (5' to 3'): CAUCCGGUAUCCCAAGACAAUCUCGGGUUGGGUUGGGAAGUAUCAUGGCUAAUC ACCAUGAUGCAAUCGGGUUGAACACUUAAUUGGGUUAAAACGGUGGGGGACGAU CCCGUAACAUCCGUCCUAACGGCGACAGACUGCACGGCCCUGCCUCAGGUGUGU CCAAUGAACAGUCGUUCCGAAAGGAAG
Puzzle: group I intron.
Crystal structure kindly provided by: BenoÎt Masquida
Reference: Mélanie Meyer, Henrik Nielsen, Vincent Oliéric, Pierre Roblin, Steinar D. Johansen, Eric Westhof and BenoÎt Masquida. 2014. Speciation of a group I intron into a lariat capping ribozyme. PNAS.111(21):7659-64.
PDB id: 4p9r
Puzzle 4
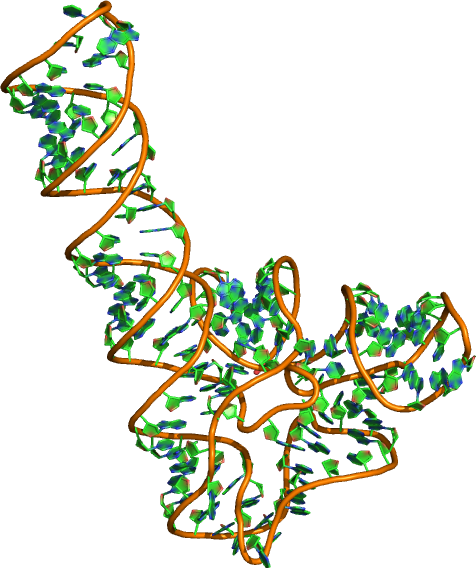
Sequence (5' to 3'): GGCUUAUCAAGAGAGGUGGAGGGACUGGCCCGAUGAAACCCGGCAACCACUAGU CUAGCGUCAGCUUCGGCUGACGCUAGGCUAGUGGUGCCAAUUCCUGCAGCGGAA ACGUUGAAAGAUGAGCCA
Puzzle: SAM-I riboswitch aptamer.
Crystal structure kindly provided by: Adrian Ferré-D'amaré
Reference: Baird NJ, Zhang J, Hamma T, Ferre-D'Amare AR. 2012. YbxF and YlxQ are bacterial homologs of L7Ae and bind K-turns but not K-loops. RNA 18, 759-770.
PDB id: 3v7e
Puzzle 3
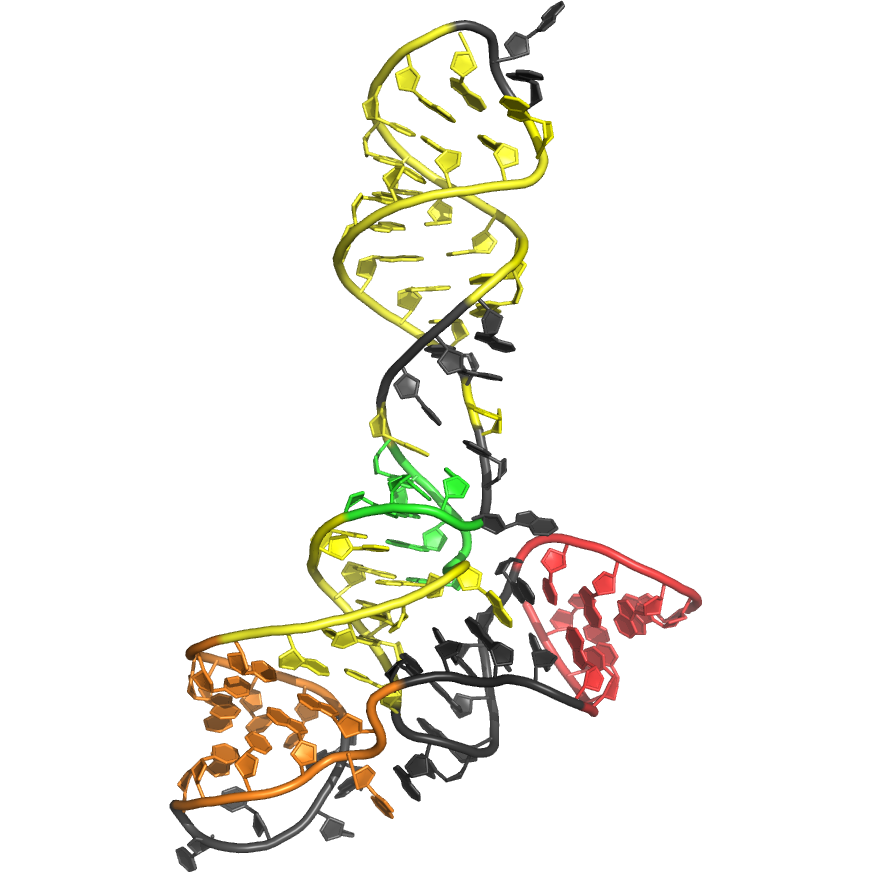
Sequence (5' to 3'): CUCUGGAGAGAACCGUUUAAUCGGUCGCCGAAGGAGCAAGCUCUGCGCAUAUGC AGAGUGAAACUCUCAGGCAAAAGGACAGAG
Puzzle: The crystallized sequence was slightly different (an apical loop was replaced by a GAAA loop) but it was not mentioned to protect the crystallographers.
Crystal structure kindly provided by: Dinshaw Patel
Reference: Huang L, Serganov A, Patel DJ. 2010. Structural Insights into Ligand Recognition by a Sensing Domain of the Cooperative Glycine Riboswitch. Mol. Cell. 40, 774-786.
PDB id: 3owz
Puzzle 2
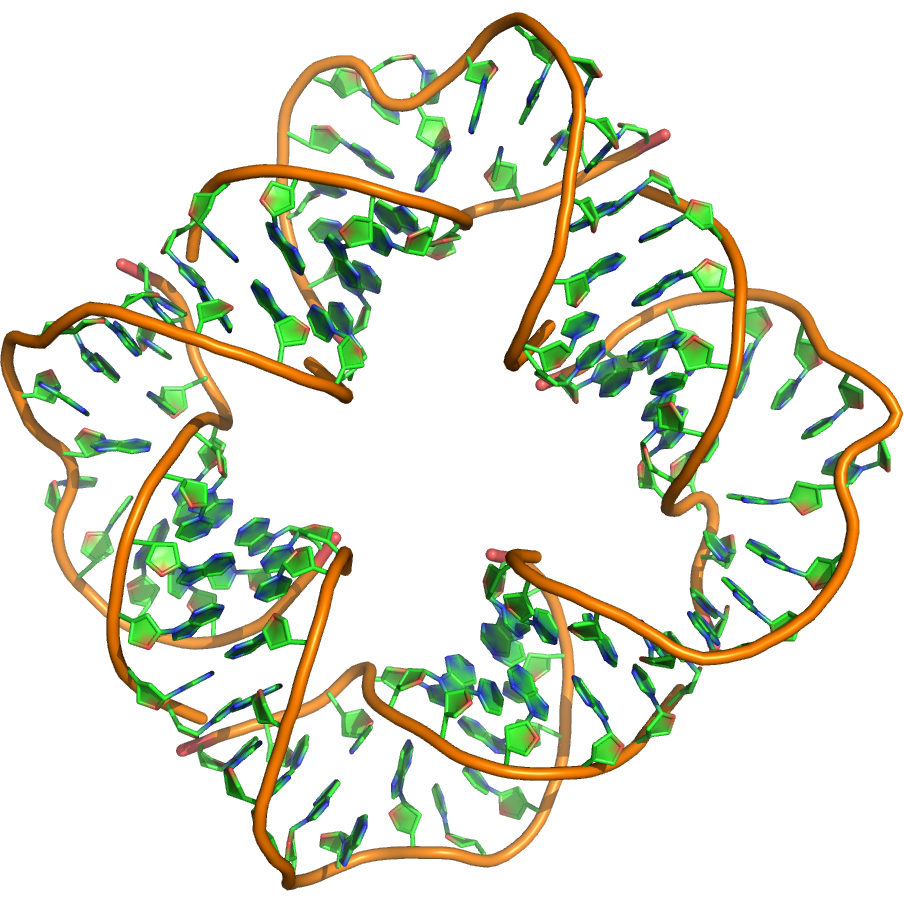
Sequence (5' to 3'):
Puzzle: The crystal structure shows a 100 nt square that assembles from four inner and four outer strands. The secondary structure shown was used for the design of the square. Actual base pairing in the crystal may deviate. 3D coordinates of the nucleotides in the inner strands (B,D,F,H) were provided. What are the structures of the outer strands (A,C,E,G)?
Crystal structure kindly provided by: Thomas Hermann
Reference: Dibrov SM, McLean J, Parsons J, Hermann T. 2011. Self-assembling RNA square. PNAS 108, 6405-6408.
PDB id: 3p59
Puzzle 1
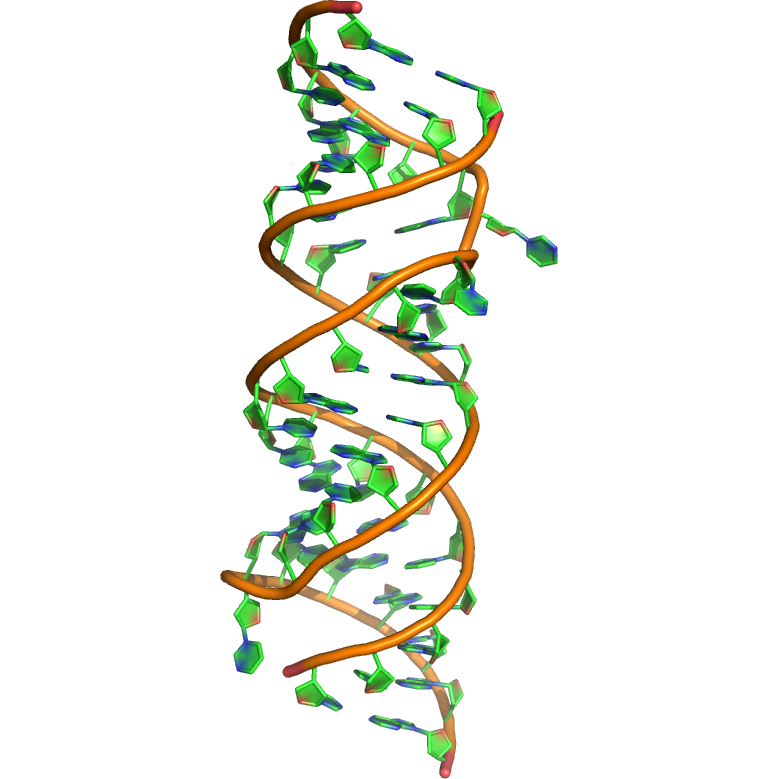
Sequence (5' to 3'): CCGCCGCGCCAUGCCUGUGGCGG
Puzzle: knowing that the crystal structure shows a homodimer that contains two strands of the sequence. The strands hybridize with blunt ends (C-G closing base pairs).
Crystal structure kindly provided by: Thomas Hermann
Reference: Dibrov SM, McLean J, Hermann T. 2011. Structure of an RNA dimer of a regulatory element from human thymidylate synthase mRNA, Acta Cryst. D, Biological Crystallography 67, 97-104.
PDB id: 3mei